Bci528Ⅰ is a Ⅱ-type restriction endonuclease from Bacillus circulans.
The strain was isolated from a soil sample and was grown in the media containing 1.5% bacto peptone, 0.5% bacto yeast extract and 0.5% NaCl at 37℃ for 18h.
The frozen cell paste was sonicated and centrifuged to take out a crude extract of Bacillus circulans 528.
Restriction endonuclease, Bci528 I was purified using following chromatographic steps: 1) Sephadex G-150 (Pharmacia), 2) DEAE-Cellulose (Whatman DE 52), 3) CB-Sepharose 6B.
Optimum conditions for Bci528Ⅰdigestion are 10mM Tris HCl, pH 7.6, 10mM MgCl2, 7mM β-mercaptoethanol and 50mM NaCl incubation at 37℃ for 1h.
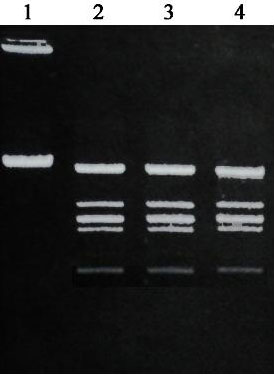
The recognition site of Bci528 I was predicted by electrophoresis analysis of digestion products by Bci528 I. The result indicates that Bci528Ⅰdigestion pattern on λ-DNA is identical to that of EcoRⅠpattern.(Fig.1) This shows that restriction enzyme Bci528Ⅰrecognizes hexanucleotide sequences 5'-GAATTC-3'
The cleavage site of Bci528Ⅰ was determined by Sanger's fluorescently labeled dideoxynucleotide chain-termination method using capillary sequencer.(Fig.2)(1) The result indicates that Bci528Ⅰ recognizes the sequence 5'-G↓AATTC-3' and cleaves between G and A of the recognition sequence producing a symmetric four base 5'overhang.
This shows Bci528Ⅰ is an isoschizomer of EcoRⅠ.(2)
The Bci528 Ⅰ enzyme has been registered the Official ReBase website (http://rebase.neb.com) under enzyme number 18749
Fig.1. Comparison of recognition sequence of Bci528Ⅰand EcoRⅠ. lane1: λ-DNA , lane2: λ-DNA+Bci528Ⅰ, lane3: λ-DNA+EcoRⅠ, lane4: λ-DNA+EcoRⅠand Bci528Ⅰ
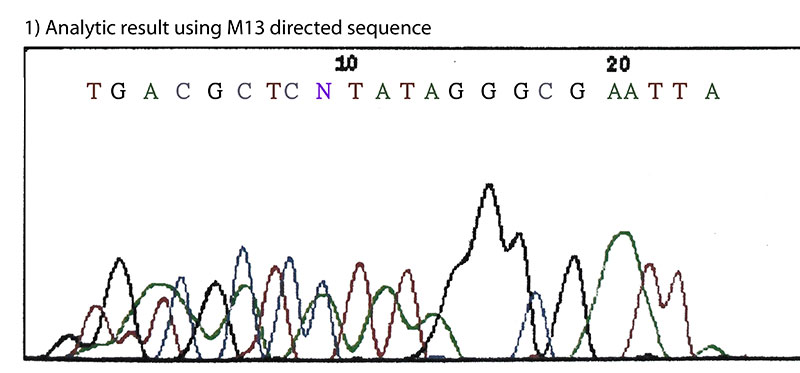
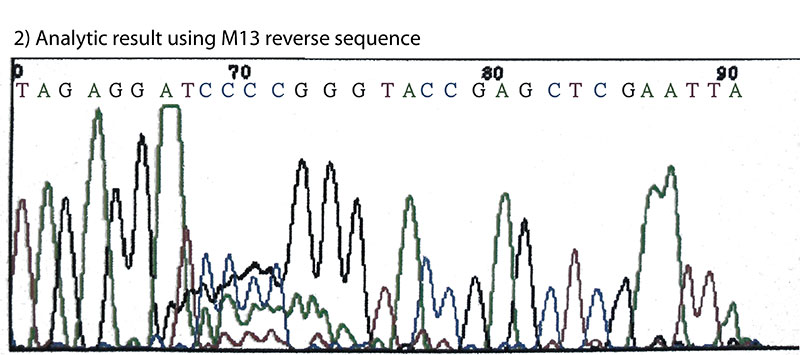
Fig.2. The cleavage site of Bci528Ⅰ.
pGEM-3Zf(+) and synthetic primer were used in sequencing reaction based upon the fluorescently labeled dideoxynucleotide chain-termination method.
M13 directed sequence: TGTAAAACGACGACGGCCAGT
M13 reverse sequence: CAGGAAACAGCTATGACC
References:
1) Sanger F. et al. (1977) Proc. Natl. Acad. Sci. USA, 74, 12, 5463.
2) Robert R.J et al. (2010) Nucl. Acids res. 38, D234.